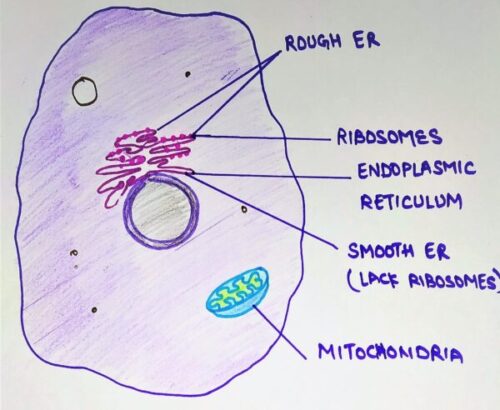
In today’s topic, we will learn about the endoplasmic reticulum and protein synthesis and segregation. The endoplasmic reticulum (ER) is an important organelle present in eukaryotic cells. It forms a network of interconnected membranous tubules or sac-like structures known as cisternae. The entire endoplasmic reticulum has a continuous membrane and it is the largest organelle of most eukaryotic cells. It plays a major role in the production, processing, and transport of proteins and lipids.
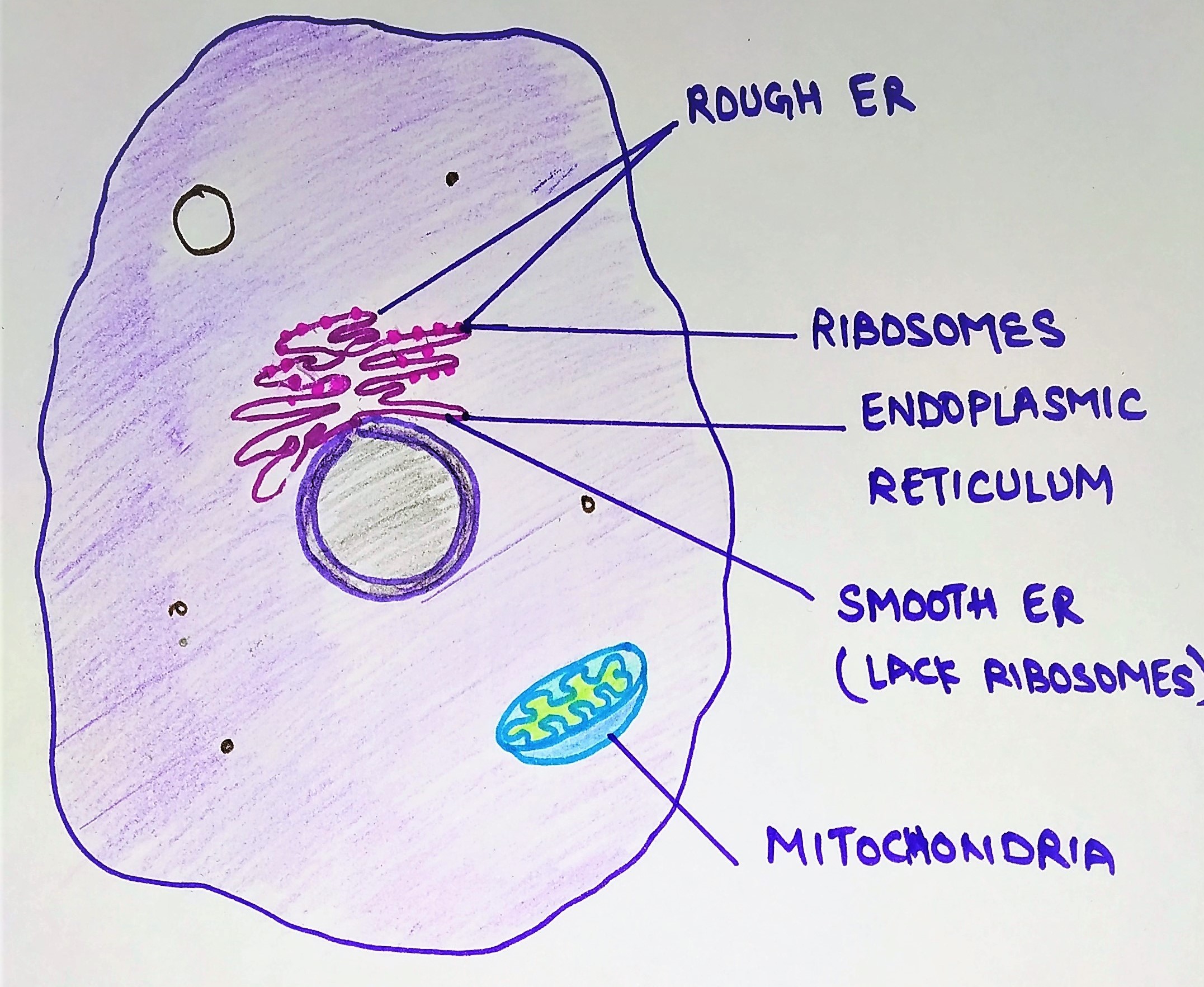
There are two regions of the ER that differ in both structure and function. Depending upon the presence or absence of ribosomes, it is of two types:
- Rough ER
- Smooth ER
Rough ER: Rough ER are cover with numerous ribosomes on their outer membrane. Usually, they are most abundant in cells that are active in protein synthesis.
Smooth ER: They lack ribosomes on their outer membrane and appear to be more vesicular with swollen cisternae. They commonly proliferate in specialized cells.
Role of endoplasmic reticulum in protein synthesis
The role of the endoplasmic reticulum in protein processing and sorting was first demonstrated by George Palade in the 1960s. He studied tissues of the exocrine pancreas.
How protein synthesis occur in endoplasmic reticulum?
Firstly, the ribosome attaches firmly to the surface of the ER membrane by its large subunit. Secondly, the ribosome binds to the rough ER by the nascent polypeptide chain.
Thirdly, the ribosomes targeted to the endoplasmic reticulum by a signal sequence at the amino terminus of the growing polypeptide chain. These signal sequences are 20 amino acids long and have short stretches of hydrophobic amino acids that are usually cleaved from the polypeptide chain during its transfer into the ER lumen. After that, signal sequence is recognized and bound by a signal recognition particle (SRP).
What is SRP?
SRP has 6 non-identical polypeptide chains and a molecule of RNA with about 300 nucleotides.
back to protein synthesis…..
An interesting aspect of SRP’s mode of action is its ability to bind to the ribosome and stop protein synthesis. The SRP does not arrest translation of all mRNAs but specifically
The docking protein has an overall basic charge on its surface, perhaps it allows its interactions with the negatively charged RNA of the SRP. Also, the docking protein binds guanosine triphosphate (GTP), which is hydrolyzed to guanosine diphosphate (GDP) during binding. The energy released by this hydrolysis fixes the SRP to the ER. At any rate, binding releases SRP, and this allows translation and translocation into the ER to proceed.
Protein destined for the insertion into the ER membrane must be prevented from following the secretory pathways. These proteins have a sequence of amino acids termed as ‘stop transfer’ signal. It acts as an anchor to keep the protein in the membrane lipid environment.
When the protein synthesis completed and detected on the surface of the rough endoplasmic reticulum, a secretory protein is located inside the RER cisternae. This step takes almost 3 minutes.
how protein translocate ?
Once the ribosome with its protein-synthesizing complex binds to the ER membrane surface via docking protein, SRP diffuses away to initiate transport of another polypeptide chain. During translocation, the attachment of ribosomes to
In order for the protein to be translocated through a membrane, it must be in an extended conformation because if it is in its native 3-dimensional globular conformation, translocation will be much more difficult. The SRP prevent the protein to adopt its 3-D conformation by acting as a molecular chaperone and keeps the protein in an extended chain.
(Those who don’t know what is a molecular chaperone, let me give you a quick brief. A molecular chaperone is a member of the family of ‘stress protein’ and their function is to stabilize non-native confirmation and to facilitate correct folding of protein subunits. Now back to our topic…..)
Thus, SRP has two roles in protein synthesis:
- It directs the complex to the ribosome and,
- It keeps the protein in the correct shape for translocation.
Soon after the signal sequence has traversed the ER membrane, a signal peptidase associated with the ER membrane cleaves off-signal sequence. Cleavage of the signal peptide appears to be an essential event for the functioning of the protein translated after it. The signal, being hydrophobic, alters the 3-D conformation of the protein existing in an aqueous environment.
Four possible modification of the protein can occur after the signal sequence cleaves.
Protein modification
- Glycosylation of protein
It is the addition of presynthesized oligosaccharides chains attached to the lipid carrier, called dolichol phosphate, which is a hydrophobic molecule embedded in the lipid bilayer of the ER membrane. The oligosaccharide chain consists of N-acetyl glucosamine, mannose and glucose, and its transfer to protein is cataly
2. Formation of disulphide bonds
These bonds are formed between cysteine residues present in the peptide chain. As a result, different regions of the protein are covalently linked, which is essential for establishing the three-dimensional structure of the protein.
3. Hydroxylation of proline and lysine residue in a polypeptide chain
This causes the interchain linkage of the fibrous protein like collagen and elastin to become stabilized. Peptidylproline hydroxylase stabilizes this reaction.
4. Carboxylation of glutamate residue
This produces the γ-glutamyl side-chain providing the modified protein with
Segregation in endoplasmic reticulum
After synthesis and modifications, protein is irreversibly segregated inside the lumen of the ER. Here irreversibly means that it should be prevented from going back to the cytoplasm through the ER membrane.
Three factors prevent it from retracing its path and these are:
- The removal of the signal sequence should stop the mature protein from interacting with the lipid bilayer since the presence of the sequence is required for binding and translocation.
- After removal of the signal sequence, the protein acquires its native 3-D globular conformation.
- Modification, say, glycosylation with amino sugars renders the protein more hydrophilic. This hydrophilic protein cannot interact with the hydrophobic membrane.
Soon after translation, the formation of multi-subunit protein occurs known as oligomerization. Most of the protein that passes through the endoplasmic reticulum is oligomeric. A molecular chaperone binds to these subunits inside the ER lumen before they acquire their globular shape and prevents them from folding inappropriately.
Role of BiP
The best example of ER chaperone is BiP, the binding protein for immunoglobulins. These proteins are made up of two types of chain and these are light and heavy. The oligomeric product has two heavy and two light chains. Once the chain is made and inserted into the ER Lumen, BiP binds to it and prevent it from aggregating. When other chains(subunits) are made and comes closer to subunit-BiP complex, BiP is released and the oligomeric product assembled.
Besides binding, BiP also plays another role and that of quality control. If a protein in the ER
In addition to “quality control”, protein degradation within the ER also appears to be an ongoing process for “quantity control”. The enzyme hydroxymethyl-glutaryl CoA reductase(HMG CoA reductase) catalyzes a key early step in the synthesis of sterols. When sterols are abundant in the cells, this enzyme degrades.
Reticuloplasmins are permanent residents of the ER lumen. These proteins have a common C-Terminal sequence. This sequence or something very similar to this sequence is a required “identity card” for a protein to be an ER-resident. For instance, deleting this sequence from the protein will cause the protein to secrete from the ER lumen and added to a protein, the protein now becomes an ER
The protein transport occurs after synthesis and segregation, from the ER to various destined locations.
Short notes
- an important organelle present in eukaryotic cells.
- forms a network of interconnected membranous tubules or sac-like structures known as cisternae.
- plays a major role in the production, processing, and transport of proteins and lipids.
Role in protein synthesis
- first demonstrated by George Palade in the 1960s on
tissues of the exocrine pancreas. - Firstly, the ribosome attaches firmly to the surface of the ER membrane by its large subunit.
- Secondly, the ribosome binds to the rough ER by the nascent polypeptide chain.
- Thirdly, the ribosomes targeted to the endoplasmic reticulum by a signal sequence at the amino terminus of the growing polypeptide chain.
- After that, the signal sequence is recognized and bound by a signal recognition particle (SRP).
- Role of SRP:
- It directs the complex to the ribosome and,
- It keeps the protein in the correct shape for translocation.
- when the protein synthesis completed and detected on the surface of the rough endoplasmic reticulum, a secretory protein is located inside the RER cisternae.
how protein translocate ?
Once the ribosome with its protein-synthesizing complex binds to the ER membrane surface, SRP diffuses away to initiate transport of another polypeptide chain.
In order for the protein to be translocated through a membrane, it must be in an extended conformation. The SRP prevent the protein to adopt its 3-D conformation by acting as a molecular chaperone and keeps the protein in an extended chain.
Soon after the signal sequence has traversed the ER membrane, a signal peptidase associated with the ER membrane cleaves off-signal sequence. Cleavage of the signal peptide appears to be an essential event for the functioning of the protein translated after it.
Four possible modification of the protein can occur after the signal sequence cleaves.
Protein modification
- Glycosylation of protein
It is the addition of presynthesized oligosaccharides chains attached to the lipid carrier, called dolichol phosphate. The oligosaccharide chain consists of N-acetyl glucosamine, mannose and glucose, and its transfer to protein is catalyzed by a group of membrane-bound enzymes called glycosyltransferases. Its attachment to protein ensures that the target protein will be in a proper 3-D configuration.
2. Formation of disulphide bonds
These bonds are formed between cysteine residues present in the peptide chain.
3. Hydroxylation of proline and lysine residue in a polypeptide chain
This causes the interchain linkage of the fibrous protein like collagen and elastin to become stabilized. Peptidylproline hydroxylase stabilizes this reaction.
4. Carboxylation of glutamate residue
This produces the γ-glutamyl side-chain providing the modified protein with
Key events in protein synthesis
- First, ribosomes attach firmly to the ER membrane by its large subunit. Almost all proteins made on the ER have a stretch of largely hydrophobic amino acids.
- The initial binding of the ribosome to the RER is mediated by the nascent polypeptide chain. This nascent polypeptide chain act as a signal with which the growing chain can attach to the membrane.
- The signal sequence translated on a free ribosome and then it binds the synthesizing complex to the ER. About 70 amino acids translated before the SRP stops it. This SRP binds the protein-synthesizing complex via a docking protein.
- Now, a signal peptidase cuts off the signal. The N-terminus is now in the ER lumen and the signal peptide cleaved. Cleavage of the signal peptide appears an essential event for the functioning of the protein translated after it.
- After cleavage, modification of proteins will occur which include glycosylation, hydroxylation, carboxylation and the formation of disulfide bonds.
- ribosomes and mRNA released just after translation completes.
Key steps in protein segregation
- After synthesis and modifications, protein irreversibly segregate inside the lumen of the ER.
- Soon after translation, the formation of multi-subunit protein occurs known as oligomerization.
- A molecular chaperone (BiP) binds to the nascent protein and prevents them from folding inappropriately.
- The completed protein bound to BiP and held in an extended shape.
- When other chains(subunits) made and come closer to subunit-BiP complex, BiP released and the oligomeric product assembled.
cytoskeleton: their structure and function
vacuoles, microbodies, and vacuoles: their structure and function
Source of information
Cell biology by David E. Sadava; CBS publisher; from page no. 213-225
Cell and molecular biology by Prakash S. Lohar; MJP Publishers; from page no. 96-98
The cell:A molecular approach by Geoffrey M. cooper, fourth edition