GOLGI COMPLEX DEFINITION
It is a membrane-bound structure found in the cytoplasm of all eukaryotic cells. If you ask me who discover the Golgi complex? My answer will be Golgi complex was discovered in the year 1898 by an Italian biologist Camillo Golgi. Hence, this important organelle got its name after him. Golgi complex is also known as the Golgi apparatus, Golgi body or you can simply call it Golgi.
You can relate this organelle to the ‘post office’. As we all know the purpose of the
GOLGI COMPLEX STRUCTURE
Thus, the Golgi complex is an organelle that sort and modifies proteins & lipids (fats) that have been built in the molecules and sends them to their destination.
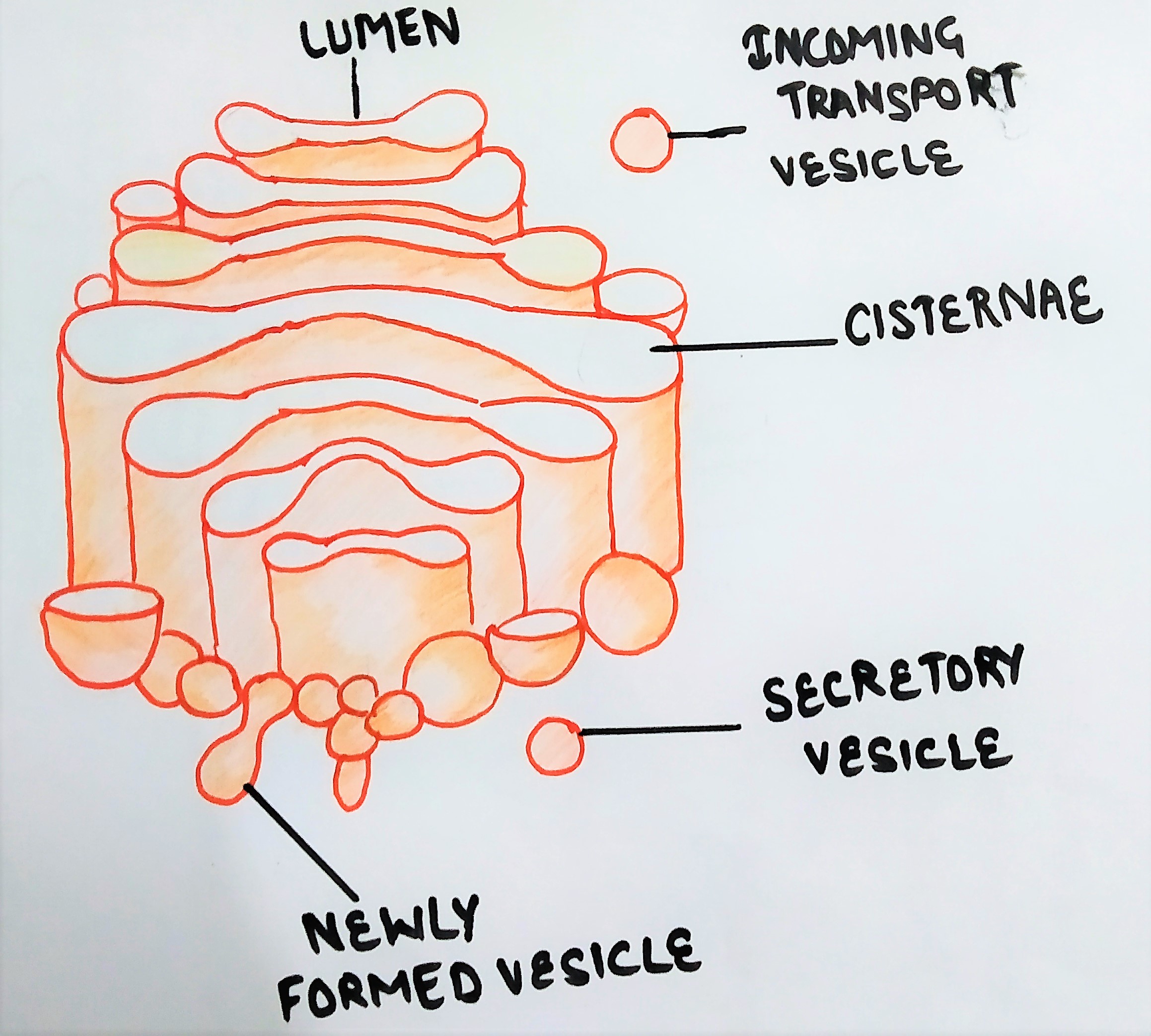
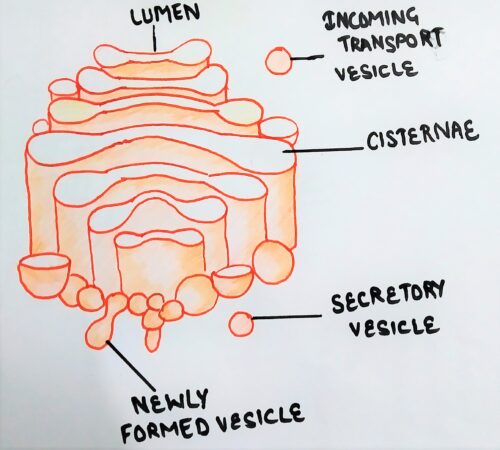
Structure of the Golgi complex includes:
- cisternae,
- tubules &
- vesicles.
Cisternae of Golgi complex
Golgi complex is made up of several fattened, stacked sacs referred to as cisternae. They are held in parallel bundles or stacks one above the other.
The Golgi cisternae on the outer face are very flat and thin whereas cisternae on the inner face or concave side are comparatively much dilated and thick. Each Golgi body consists of 4-8 cisternae, slightly curved, each one has a diameter of 0.5-1.0mm.
The membrane of cisternae is smooth and fenestrated (porous) and the pores may be localized or present all over the surface.
Cisternae of the Golgi complex has three regions
- cis face
- trans face.
- Medial region
Cis face
The ‘cis’ face is convex in appearance and it faces the nucleus and endoplasmic reticulum and has smaller vesicles of 50 nm in diameter. It acts as a
Trans face
The’ trans’ region faces the plasma membrane and has larger vesicles associated with it. The vesicles that leave the Golgi apparatus exit from its trans face.
medial face
In between ‘cis‘ and ‘trans’ cisternae, the stack of cisternae is called medial cisternae.
Golgi Tubules
The tubules arise from the peripheral region of cisternae. Some cisternae have 60 nm diameter tubules. Further, these tubules are usually connected with the ER or adjacent cisternae.
Golgi Vesicles
The vesicles are goblet- like structures attached to the tubules. Both coated and uncoated vesicles are associated with Golgi.
The term dictyosome refers to individual stacks in plant cells. The no. of stacks depends on cell type and physiological function. For example, certain cells in fungi have a single cisterna; hepatocytes have about 50 stacks; pollen tubes of higher plants have as many as 25,000 stacks.
ROLE OF GOLGI COMPLEX IN PROTEIN SECRETION
Transport from ER to Golgi
Golgi membranes have an obvious transition in composition between the Endoplasmic Reticulum and the plasma membrane.
Proteins move between the compartments in vesicles that bud off from one compartment and then fuse with the next. The vesicles are coated with a spike protein which is totally unrelated to clathrin.
Guanosine triphosphate ( GTP) hydrolysis is required for the transport of the vesicle at the
Modification of secretory proteins
Secretory proteins are synthesized and processed by the RER, where modifications occur. In the Golgi complex, a no. of additional covalent modifications can occur. The most common of these is glycosylation. The carbohydrates are added in sequence, in separate compartments of the Golgi complex, as the protein passes through the cisternae.
Each reaction in the Golgi changes the protein to make it inaccessible to the enzyme for the next step.
Other modifications of secretory proteins are sulfation and proteolysis.
Movement through the Golgi complex
Molecules move within the Golgi complex by several mechanisms:
- by the migration of entire cisternae:
the migration of entire cisternae from one face towards the other is accompanied by a loss of trans vesicles. More cis cisternae would be added via vesicles from the ER and the plasma membrane is recycled by endocytosis.
- By interconnections between the cisternae
Golgi is a static structure. Transfer of protein would be through interconnections between the cisternae, in a vectoral fashion.
- Comparison between the other two
The cisternae bud off to form other vesicles, which shuttle from one cisterna to the next. Vesicle that fuses with the plasma membrane would essentially be returned to the Golgi via endocytosis.
Packaging of secretory molecules.
After modifications, first, the secretory proteins often appear in amorphous condensing vacuoles. After that, they appear in more concentrated secretory granules.
Electron microscopy indicates that the granules membranes come from the Golgi membrane. The
Electrostatic interactions between cationic and anionic secretion products could form complexes that reduce the effective osmotic concentration in the vacuole. After that, Water would move out into the cytoplasm. As a result, vacuole concentrate to a granular appearance.
Generally, the contents in the secretory granules are a “mixed bag”, with each granule having a random assortment of all of the molecules that need to be secreted.
All molecules that need to be secreted are packed by Golgi, but what will happen to the molecules that are not secreted by the organelle. Want to know? Then move ahead…..
Golgi complex is also responsible for modifications of non-secreted proteins. These proteins are the ones that are bound for the
Examples of such proteins
- The viral G-protein: the viral G-protein is co-translationally glycosylated in the RER by the addition of glucosamine and
polymannose . In the cis-Golgi element, mannose residues are removed, and in the trans-Golgi, capping sugars are added. - Lysosomal enzymes: enzymes destined for the lysosomes are mainly hydrolases. Hydrolases are glycosylated on the RER. In the cis Golgi cisternae, a mannosidase removes some mannose residues. In the medial Golgi, N- acetylglucosamine
phosophotransferase puts N-acetyl-glucoseamine -1-phosphate on the mannose residues. A glucosaminidase removes the N-acetylglucosamine. In the end, it leaves mannose phosphate as the terminal sugar on lysosomal hydrolases.
Sorting
There must be a mechanism to direct the secretory protein to their final destinations in the cell.
Sorting of lysosomal hydrolases
The recognition factor on the extracellular lysosomal hydrolases is
The two enzymes involved in phosphomannose production are located in the cis and medial elements of the Golgi complex. The modified enzymes are then moves to the trans region, where sorting occurs.
Sorting mechanism
The actual sorting mechanism is a receptor for the phosphomannose-coated protein. This receptor is a membrane protein located in the trans Golgi elements and binds the Golgi-modified protein via their mannose phosphate groups.
Binding is best at slightly acidic pH; if the pH falls below 5.5, the ligand and receptor dissociate.
firstly, the membrane with its receptor attached to the lysosomal enzyme. After that, it buds off the Golgi as a clathrin-coated vesicle. The proton pump will gradually acidify the vesicle. As a result, the enzyme dissociates from the receptor. The enzyme will go to the lysosomes, while the receptor
Note: the phosphomannose recognition system is apparently not operative in all tissues. Certain cells contain the enzymes for the
Now, wonder…… what happens to the protein that resides in the Golgi?
Another sorting function of the Golgi is to target the proteins that are resident in the ER. These proteins usually have the KDEL terminal amino acid signal which usually
How protein resides in the Golgi?
If you are thinking that how a protein will reside in Golgi, for such
There must be a signal for the retention of Golgi-resident proteins, such as the enzymes catalyzing protein modifications. It appears that some part of the protein causes it to remain in the Golgi, but there must be a specific sequence of amino acids required for the same. For example, the protein EI coded by the infectious bronchitis virus localized in the cis Golgi and has three membrane-spanning sequences. The first of these is apparently for Golgi retention. If it is missing, the protein will secrete.
Golgi-resident proteins are all membrane proteins and there appear to be no soluble proteins that reside in the Golgi.
It seems that there is no signal for the targeting of proteins from the Golgi to the plasma membrane or the extracellular medium. Instead, secretion is the default pathway, which occurs when other signals not detected at the Golgi.
BIOGENESIS OF GOLGI COMPLEX
As I told you before that Golgi is an essential membrane-bound organelle located in the eukaryotic cells.
In mammalian cells, the Golgi stacks integrate into a continuous perinuclear ribbon, which poses a challenge for the daughter cells to inherit this membrane organelle during cell division.
In mammalian cells, the continuous Golgi ribbon should segregate into the two daughter cells when the cell divides. This is achieved through a three-step process
- Disassembly of Golgi membrane,
- Partitioning, &
- Reassembly of Golgi membrane.
Disassembly
In the late G2 phase of the cell cycle, the Golgi ribbon will unlink upon separation of the lateral connections between the stacks.
The conversion of the ribbon into stacks depends on the MAP kinase, kinase MEK1. Golgi ribbon unlinking is caused by kinase Mek1 through mitotic phosphorylation.
(Kinases are enzymes that catalyses the transfer of a phosphate group from ATP to a specified molecule.)
Besides these kinases, the membrane fission protein CtBP1/BARS is important for Golgi ribbon unlinking.
Inhibition of BARS activity prevents severing (break off) of the ribbon as well as G2/M transition.
Partitioning
After the conversion of the Golgi ribbon into stacks, the cisternae unstacked and vesiculated. Vesiculation of the Golgi cisternae is triggered by an imbalance of membrane budding and fusion during mitosis.
In interphase, COPI vesicles captured and tethered to the cis-cisternae by the ternary giantin-p115-GM130 tethering complex. Once the complex formed, p115 directly catalysis the formation of SNARE complexes (SNARE proteins mediate vesicle fusion), leading to the
During mitosis, COPI vesicles continue to form, but the vesicles cannot fuse with their target membranes because of the disruption of vesicle tethering complex.
The persistent budding of COPI vesicles without fusion gradually consumes the cisternae.
Efficient disassembly of the Golgi in early mitosis facilitated by the
Reassembly
Upon segregation into the nascent daughter cells, the mitotic Golgi membranes reassemble during telophase and cytokinesis into two distinct ribbons on opposite sides of the nucleus.
A smaller Golgi ribbon reforms adjacent to the midbody, whereas a larger ribbon found in the pericentriolar region on the side away from the cleavage furrow.
Following abscission, the smaller ribbon moves to the opposite side of the nucleus and merges with the larger Golgi into a continuous ribbon.
the Golgi reformed at the end of mitosis by two interrelated processes,
- formation of flattened cisternae by membrane fusion and
- stacking of cisternae.
Following segregation into the daughter cells, a functional Golgi is reformed.
SHORT NOTES
Golgi complex
- membrane-bound structure found in the cytoplasm of all eukaryotic cells.
- discovered in the year 1898 by an Italian biologist Camillo Golgi.
- also known as the Golgi apparatus, Golgi body or Golgi.
- sorts and modifies proteins & lipids.
structure
- Structure of the Golgi complex includes:
- cisternae
- cis face
- trans face
- medial face
- tubules
- vesicles
- cisternae
cisternae of Golgi complex
- each Golgi body consists of 4-8 cisternae, each one has a diameter of 0.5-1.0mm.
- made up of several fattened, stacked sacs.
- they are held in parallel bundles or stacks one above the other.
- very flat and thin on the outer face whereas dilated and thick at the inner face.
- the membrane of cisternae is smooth and porous and the pores may be localized or present all over the surface.
Cis face of cisternae
- convex in appearance and faces the nucleus and ER.
- act as the receiving compartment from
ER . - the vesicle that
leave the ER fuse to thegolgi from here.
Trans face
- faces the plasma membrane and has larger vesicles.
- vesicles that leave the Golgi apparatus exit from its trans face.
medial face
- present between
the ‘cis ‘ and ‘trans’ cisternae.
Golgi Tubules
- arise from the peripheral region of cisternae.
- they are usually connected with the ER or adjacent cisternae.
- some cisternae have 60 nm diameter tubules.
Golgi Vesicles
- goblet- like structures attached to the tubules.
- Both coated and uncoated vesicles are associated with Golgi.
The term dictyosome refers to individual stacks in plant cells. In addition, The no. of stacks depends on cell type and physiological function. For example, certain cells in fungi have a single cisterna; hepatocytes have about 50 stacks; pollen tubes of higher plants have as many as 25,000 stacks.
Role of Golgi complex in protein secretion
Transport from ER to Golgi
- proteins move between the compartments in vesicles that bud off from one compartment and after that, fuse with the next.
- the vesicles are coated with a spike protein. Guanosine triphosphate ( GTP) hydrolysis is required for the transport of the vesicle at the endoplasmic reticulum, where it loses its coating.
Modification of secretory proteins
- modifications occur at RER.
- A no. of additional covalent modifications also occur in the Golgi complex.
- glycosylation: carbohydrates are added in sequence as the protein passes through the cisternae.
- each reaction in the Golgi changes the protein to make it inaccessible to the enzyme for the next step.
- other modifications of secretory proteins are sulfation and proteolysis.
Movement through the Golgi complex
Molecules move within the Golgi complex by several mechanisms:
- by
migration of entire cisternae:it is accompanied by a loss of trans vesicles. More cis cisternae would be added via vesicles from the ER and the plasma membrane is recycled by endocytosis.
- By interconnections between the cisternae: transfer of protein would be through interconnections between the cisternae, in a vectorial fashion.
- Comparison between the other two: the cisternae bud off to form other vesicles. In addition, vesicle that fuses with the plasma membrane would essentially returned to the Golgi via endocytosis.
Packaging of secretory molecules
- first, the secretory proteins appear in amorphous condensing vacuoles. After that, they appear in more concentrated secretory granules.
- the Concentration of the vesicle contents occurs in the presence of uncouplers of oxidative phosphorylation. As a result, this step does not require the continuous presence of ATP.
- in the vacuole,
effective osmotic concentration will reduced. As a result, vacuole concentrate to a granular appearance.
Golgi complex is also responsible for modifications of non-secreted proteins.
Examples of such proteins
- The viral G-protein: In the cis-Golgi element, mannose residues will be removed, and in the trans-Golgi, capping sugars will be added.
- Lysosomal enzymes: hydrolases are glycosylated on the RER. In the cis Golgi cisternae, a mannosidase removes some mannose residues. In the medial Golgi, N- acetylglucosamine
phosophotransferase puts N-acetyl-glucoseamine -1-phosphate on the mannose residues. A glucosaminidase removes the N-acetylglucosamine. In the end,
Sorting
There must be a mechanism to direct the secretory protein to their final destinations in the cell.
Sorting of lysosomal hydrolases
- the recognition factor is
phosphomannose . In this case, sorting occurs in the trans Golgi elements. - the two enzymes involved in
phosphomannose production are located in the cis and medial elements of the Golgi complex. The modified enzymesare then moves to the trans region, where sorting occurs.
Sorting mechanism
- The actual sorting mechanism is a receptor for the phosphomannose-coated protein. This receptor is a membrane protein located in the trans-Golgi elements and binds the Golgi-modified protein via their mannose phosphate groups.
- Binding is best at slightly acidic pH; if the pH falls below 5.5, the ligand and receptor dissociate.
- firstly, the membrane with its receptor attached to the lysosomal enzyme. After that, it buds off the Golgi as a clathrin-coated vesicle.
- The proton pump will gradually acidify the vesicle. As a result, the enzyme dissociates from the receptor.
- In the end, the enzyme will go to the lysosomes, while the receptor will recycle back to the trans-Golgi, where it can pick up another lysosome-targeted enzyme.
How protein reside in the Golgi?
- There must be a signal for the retention of Golgi-resident proteins, such as the enzymes catalyzing protein modifications. It appears that some part of the protein causes it to remain in the Golgi, but there must be a specific sequence of amino acids required for the same. For example, the protein EI coded by the infectious bronchitis virus localized in the cis Golgi and has three membrane-spanning sequences. The first of these is apparently for Golgi retention. If it is missing, the protein will secrete.
Golgi Biogenesis
In mammalian cells, the continuous Golgi ribbon should segregate into the two daughter cells when the cell divides. This is achieved through a three-step process
- Disassembly of Golgi membrane
- Partitioning &
- Reassembly of Golgi membrane
Disassembly of Golgi membrane
- In the late G2 phase of the cell cycle, the Golgi ribbon will unlink.
- Golgi ribbon unlinking is caused by kinase Mek1 through mitotic phosphorylation.
- Besides these kinases, the membrane fission protein CtBP1/BARS is important for Golgi ribbon unlinking.
- Inhibition of BARS activity prevents severing (break off) of the ribbon as well as G2/M transition.
Partitioning
- After the conversion of the Golgi ribbon into stacks, the cisternae unstacked and vesiculated. this step is triggered by an imbalance of membrane budding and fusion during mitosis.
- In interphase, COPI vesicles captured and tethered to the cis-cisternae by the ternary giantin-p115-GM130 tethering complex.
- after that, p115 directly catalysis the formation of SNARE complexes, leading to the fusion of the two membranes.
- During mitosis, COPI vesicles continue to form, but the vesicles cannot fuse with their target membranes because of the disruption of vesicle tethering complex.
Reassembly
- upon segregation into the nascent daughter cells, the mitotic Golgi membranes reassemble during telophase and cytokinesis into two distinct ribbons on opposite sides of the nucleus.
- a smaller Golgi ribbon reformed adjacent to the midbody. On the other hand, a larger ribbon found in the pericentriolar region.
- following abscission, the smaller ribbon moves to the opposite side of the nucleus and merges with the larger Golgi into a continuous ribbon.
- the Golgi reformed at the end of mitosis by two interrelated processes,
formation of flattened cisternae by membrane fusion and- stacking of cisternae.
- following segregation into the daughter cells, a functional Golgi is reformed.
REFERENCES
Cell biology: organelle structure and function by David E. Sadava. Page no.241-263
Cell and Molecular biology by Prakash S. Lohar, Golgi complex
https://academic.oup.com/jb/article-abstract/137/2/115/873179?redirectedFrom=fulltext